About Me
Hello and welcome to my personal website!I'm Mengqi Wang, a passionate PhD student in Animal Science, specializing in the fascinating fields of genetics and epigenetics, with a particular emphasis on unraveling the intricacies of DNA methylation.
Throughout my academic journey,
I have been fascinated by the complexity of epigenetic mechanisms and their crucial roles in regulating gene expression and phenotypic variation.
My resaech revolves around investigating the epigenetic mechanisms, including DNA methylation and non-coding RNA, involved in the reulation of important
biological processes in animals, such as the milk production and mastitis of dairy cows, and their potential applications in breeding programs and therapeutic interventions.
Moreover, I am also interested in exploring the regulatory roles and potential applications of epigenetics in human health,
particularly in complex diseases such as cancers.
Through my research, I have acquired extensive experience in laboratory techniques, bioinformatics, and data analysis.
I have published several research articles in international peer-reviewed journals, and my work has been presented at various national and international conferences.
As I am about to complete my PhD study,
I am excited to seek new opportunities to expand my research horizons and collaborate with experts in the field.
Therefore, I am actively seeking job offers such as postdoctoral positions where I can apply my skills and knowledge to advance the field of epigenetics in human and animal health.
Thank you for visiting my website. Please feel free to browse through my research interests, publications,
and academic background. I am looking forward to hearing from you and discussing potential collaborations.

Research Experience
Throughout my academic career, I have been fortunate to engage in diverse and impactful research experiences, delving into the intricacies of animal genetics and epigenetics. These research endeavors have allowed me to contribute to the advancement of knowledge in my field and have shaped my passion for unraveling the complexities of genetics/epigentics and its implications for animal health and productivity.Jan. 2020 - Sep. 2023
- Project Participation: Roles of DNA methylation marks in the modulation of subclinical mastitis in Canadian Holstein cows
- Responsibility: Sample collection and processing, wet-lab, bioinformatics analysis for RNA sequencing and DNA methylation sequencing data, including whole-genome bisulfite sequencing and targeted amplicon bisulfite sequencing, and manuscript writing.
Sep. 2016 - Jun. 2019
- Project Participation: The molecular regulatory mechanisms of DNA methylation and miRNA underlying bovine mastitis induced by S. aureus
- Responsibility: Sample collection and processing, DNA methylation seuqnencing data analysis, gene polymorphism detection, cell culture, molecular experiments for verifying the function of microRNAs, and manuscript writing.
Apr. 2018 - Apr. 2019
- Project Participation: Roles of non-coding RNA in Johne's disease progression in gastrointestinal tract of dairy cows
- Responsibility: Sample collection and processing, wet-lab (DNA isolation, PCR, qRT-PCR), data analysis, and manuscript writing.
Sept. 2014 - Jun. 2016
- Project Participation: Effects of three grass combination on nitrate reduction in rumen fluid of Hu sheep
- Responsibility: Rumen content collection and processing, culture of rumen bacteria, analyze nitrate metabolism in rumen, bacteria DNA extraction, and manuscript writing.
Education Experience
I have a strong educational background in the field of animal science, equipping me with a solid understanding of the complex genetics and epigenetics.Jan. 2020 - Sep. 2023
Laval University, Quebec, Quebec, Canada.
Cooperate with Agriculture and Agri-Food Canada - Sherbrooke Research and Development Center, Sherbrooke, Canada.
Thesis title: Regulatory roles of DNA methylation and its potential as epigenetic marker in bovine subclinical mastitis.
Sep. 2016 - Jun. 2019
Yangzhou University, Yangzhou, Jiangsu, China
Cooperate with Agriculture and Agri-Food Canada - Sherbrooke Research and Development Center, Sherbrooke, Canada.
Thesis title: Genome-wide DNA methylation analysis of mammary gland tissues from Chinese Holstein cows with Staphylococcus aureus induced mastitis.
Sep. 2012 - Jun. 2016
Yangzhou University, Yangzhou, Jiangsu, China
Thesis title: Effects of three grass combination on the nitrate reduction in rumen fluid of Hu Sheep
What I’m good at?
Strong expertise in laboratory techniques, bioinformatics analysis, and data interpretation; excellent scientific writing and communication skills; adept at project management and animal handling for genetic and epigenetic research.Scientific Communication
A commendable aptitude for both written and oral communication, demonstrated through the expertise in presenting research findings at numerous international conferences and producing peer-reviewed publications.
Laboratory Techniques
Proficient in various molecular biology techniques, including DNA/RNA extraction, PCR amplification, RT-qPCR, gel electrophoresis, and etc.. Skilled in functional studying about DNA methylation and non-coding RNAs.
Bioinformatics
Experienced in analyzing next-generation sequencing data, including RNA sequencing, small RNA sequencing and whole genome DNA methylation sequencing data, utilizing bioinformatics tools, and applying multi-Omics approaches for revealing the complexity of diseases.
Data Analysis
Good at statistical analysis and utilizing programming languages such as R and Python for data manipulation and analysis. Proficient in data visualization by using R, Google Charts, Apache ECharts, GraphPad Prism, Adobe Photoshop and Adobe Illustrator.
Research Interests
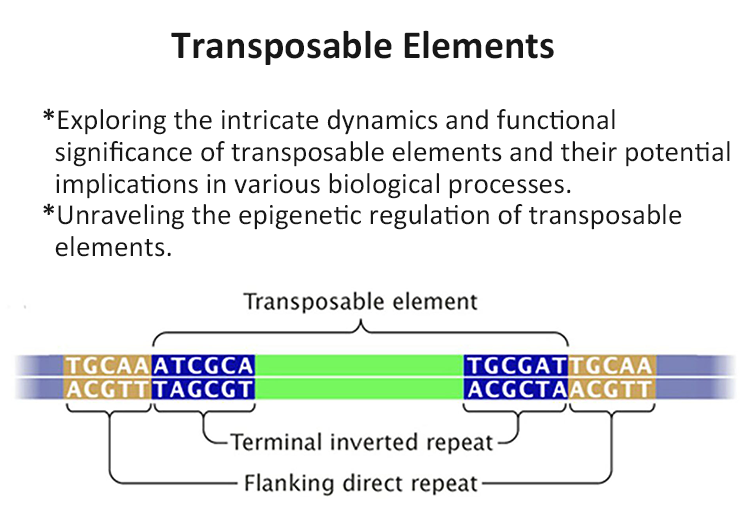
In my research endeavors, I am passionate about exploring the intricate interplay between genetics and epigenetics underlying diseases. I am particularly interested in investigating the role of DNA methylation in shaping phenotypic traits, unraveling the underlying mechanisms, and understanding its implications for health and productivity.
Awards and Hornors
Acknowledgment of my dedication and academic achievements has been bestowed through various awards and honors throughout my academic journey. These recognitions serve as a testament to my commitment to excellence in the field of Animal Genetics and Epigenetics. I am grateful for the opportunities and recognition that have come my way, fueling my passion for advancing scientific knowledge and making a meaningful impact in the research community.
- Congress travel scholarship from Op+lait to attend Plant & Animal Genome Conference (San Diego, USA, 2023)
- Travel fellowship for PhD students to attend World Congress on Genetics Applied to Livestock Production Meeting (Rotterdam, Netherlands, 2022)
- Best poster presentation at 2022 Symposium sur les bovins laitiers (Quebec, Canada, 2022)
- Third place Oral Competition winner, CSAS Graduate Student competition, 2021 ASAS-CSAS Annual Meeting & Trade Show (Louisville, USA, 2021)
- China Scholarship Council (CSC) Scholarship. Impact: Provided full tuition coverage and generous stipend for PhD study, facilitating focused research and academic growth. Significance: Highly competitive scholarship showcasing academic excellence and fostering international academic exchange. (2020-2023)
- China Scholarship Council (CSC) Scholarship. Impact: Provided generous stipend for studing in Agriculture and Agri-Food Canada as a student intern. (April, 2018 - April, 2019)
- China National Scholarship for graduate student (2018)
- The 2nd Dairy Opportunity & Challen Competition of National agricultural and Forestry Universities, China, First Place Honor (2017)
- China National Scholarship for graduate student (2017)
- China National Scholarship for Encouragement (2015)
- China National Scholarship for undergrate student (2014)
- China National Scholarship for Encouragement (2013)
Publications
During my academic journey, I have had the privilege of contributing to the scientific community through the publication of several research articles in peer-reviewed journals. These publications reflect my dedication to rigorous research, innovative findings, and the dissemination of knowledge in my field.Research articles in international peer-reviewed journals:
- Wang, Mengqi, Nathalie Bissonnette, Mario Laterrière, Pier-Luc Dudemaine, David Gagné, Jean-Philippe Roy, Marc-André Sirard, and Eveline M. Ibeagha-Awemu. "Genome-wide DNA methylation and transcriptome integration associates DNA methylation changes with bovine subclinical mastitis caused by Staphylococcus chromogenes." International Journal of Molecular Sciences 24, no. 12 (2023):10369.
- Wang, Mengqi, Nathalie Bissonnette, Mario Laterrière, Pier-Luc Dudemaine, David Gagné, Jean-Philippe Roy, Marc-André Sirard, and Eveline M. Ibeagha-Awemu. "Gene co-expression in response to Staphylococcus aureus infection reveals networks of genes with specific functions during bovine subclinical mastitis." Journal of Dairy Science. 106, no. 8, (2023):5517-5536.
- Wang, Mengqi, Nathalie Bissonnette, Mario Laterrière, Pier-Luc Dudemaine, David Gagné, Jean-Philippe Roy, Xin Zhao, Marc-André Sirard, and Eveline M. Ibeagha-Awemu. "Methylome and transcriptome data integration reveals potential roles of DNA methylation and candidate biomarkers of cow Streptococcus uberis subclinical mastitis." Journal of Animal Science and Biotechnology 13, no. 1 (2022): 136.
- Wang, Mengqi, Nathalie Bissonnette, Pier-Luc Dudemaine, Xin Zhao, and Eveline M. Ibeagha-Awemu. "Whole genome DNA methylation variations in mammary gland tissues from holstein cattle producing milk with various fat and protein contents." Genes 12, no. 11 (2021): 1727.
- Wang, Mengqi, and Eveline M. Ibeagha-Awemu. "Impacts of epigenetic processes on the health and productivity of livestock." Frontiers in Genetics 11 (2021): 613636.
- Wang, Mengqi, Yan Liang, Eveline M. Ibeagha-Awemu, Mingxun Li, Huimin Zhang, Zhi Chen, Yujia Sun, Niel A. Karrow, Zhangping Yang, and Yongjiang Mao. "Genome-wide DNA methylation analysis of mammary gland tissues from Chinese Holstein cows with Staphylococcus aureus induced mastitis." Frontiers in Genetics 11 (2020): 550515.
- Wang, Mengqi, Duy N. Do, Camille Peignier, Pier-Luc Dudemaine, Flavio S. Schenkel, Filippo Miglior, Xin Zhao, and Eveline M. Ibeagha-Awemu. "Cholesterol deficiency haplotype frequency and its impact on milk production and milk cholesterol content in Canadian Holstein cows." Canadian Journal of Animal Science 100, no. 4 (2020): 786-791.
- Wang, Mengqi, Hailiang Song, Xiaorui Zhu, Shiyu Xing, Meirong Zhang, Huimin Zhang, Xiaolong Wang, Zhangping Yang, Xiangdong Ding, Niel A. Karrow, Sven König and Yongjiang Mao. "Toll-like receptor 4 gene polymorphisms influence milk production traits in Chinese Holstein cows." Journal of Dairy Research 85, no. 4 (2018): 407-411.
- Wang, Mengqi, Wei Ni, Huimin Zhang, Xiaolong Wang, Zhangping Yang, Eveline M. Ibeagha-Awemu, and Yongjiang Mao. "Parity-dependent association between IL-12Rβ2 and IL-23R gene polymorphisms and clinical mastitis in Chinese Holstein cows." Indian Journal of Animal Research 52, no. 12 (2018): 1705-1710.
Research articles submitted international peer-reviewed journals or in preparation:
- Wang, Mengqi, Nathalie Bissonnette, Mario Laterrière, Pier-Luc Dudemaine, David Gagné, Jean-Philippe Roy, Marc-André Sirard, and Eveline M. Ibeagha-Awemu. "DNA methylation haplotype block signatures responding to S. aureus subclinical mastitis and association with production and health traits" Submitted to BMC Biology. Under review.
- Wang, Mengqi, et al. "Multi-omics data integration reveals the genes, non-coding RNAs and DNA methylation network and hub signatures regulating bovine subclinical mastitis." Submitted to Frontiers in Genetics.
- Wang, Mengqi, et al. "MicroRNA potential regulatory functions in cow’s ileum and ileum lymph node response to Mycobacterium avium subspecies paratuberculosis infection." (In preparation).
- Wang, Mengqi, et al. "MicroRNA profiles in jejunum and jejunal lymph node and their potential regulatory roles in response to Johne’s disease infected by Mycobacterium avium subspecies paratuberculosis" (In preparation).
Research articles in Chinese peer-reviewed journals:
- Wang MQ, Ni W, Tang C, et al. Association Analysis on the SNP of LF -131 C>T and LF -28 A>C with Milk Performance, Clinical Mastitis and Lifetime for Chinese Holstein (Bos taurus). Journal of Agricultural Biotechnology, 2018, 26(5): 811-818.
- Wang MQ, Ni W, Xia HL, et al. The Analysis of Factors Affecting the Gestation length of Chinese Holstein. Animal Husbandry and Veterinary Medicine, 2018,50(3):1-5.
- Wang MQ, Ni W, Tang C, et al. Effects of calving seasons on growth curve fitting of Holstein cattle. Journal of Southern Agriculture, 2018, 49(11).
- Wang MQ, NI W, Tang C, et al. Comparative Analysis of Milk Production Traits between Jersey and Holstein. Journal of Domestic Animal Ecology, 2018,39(7): 37-41.
- Wang MQ, Xing SY, Ni W, et al. Association Analysis Between SNPs of CXCR1 Gene and Susceptibility to Bovine Leukemia in Chinese Holstein (Bos taurus). Journal of Agricultural Biotechnology, 2017, 25(10): 1637-1642.
- Wang MQ, NI W, Tang C, et al. Association analysis between polymorphism of CXCR1 gene coding region and milk production and partial reproductive traits in Holstein cattle. Journal of Northeast Agricultural University, 2017, 48(11):35-42.
- Wang MQ, Zhu XR, Xing SY, et al. Effect of FADS2 c.908 C>T mutation on milk production traits and fatty acids composition in milk of Chinese Holstein. Journal of Northwest A&F University, 2017, 45(3):1-6.
- Wang MQ, Ni W, Zhang HM, et al. Correlation Between the Mutation of SNPs in the Promoter Region of TLR1 and Mastitis Resistance and Milking Traits in Chinese Holstein. Journal of Agricultural Biotechnology, 2017, 25(3): 397-404.
- Wang MQ, Ni W, Zhang HM, et al. Association Between SNPs in the CDS Regions of CXCR1 Gene and the Clinical Mastitis and Lifetime for Chinese Holstein. Scientia Agricultura Sinica, 2017, 50(12):2359-2370.
- Wang MQ, NI W, Guo YW, et al. Correlation Analysis between the Abortion, Diseases and Milking Traits and SNP of SLC35A3-66 for Chinese Holstein Cows. China Journal of Veterinary Science, 2017, 37(12):2418-2424.
- Xun CX, Wang MQ, Zhu XR, et al. Effects on the composition of fatty acids in milk of SNPs in the 3’ untranslated regions of FADS2 for Chinese Holstein. Scientia Agricultura Sinica, 2016,49 (11): 2194-2202, Co-first author.
- Wang MQ, Zhu XR, Xing SY, et al. Factors Affecting the birth weight of Holstein calf. Journal of Domestic Animal Ecology, 2016,37 (11):22-25.
- Wang MQ, Chen ZY, Ma TT, Lin Miao. Effects of three grass combination on the nitrate reduction in rumen fluid of Hu sheep. Pratacultural Science, 2016, 33 (3): 489-494.
Outreach
In addition to my publications, I have had the opportunity to showcase my research findings at numerous national and international conferences. These presentations have allowed me to engage with fellow researchers, receive valuable feedback, and contribute to the exchange of ideas in my field.Oral presentations:
- Mengqi Wang, Nathalie Bissonnette, Mario Laterrière, David Gagné, Marc-André Sirard, and Eveline M. Ibeagha-Awemu. DNA methylation of first exons negatively correlate with gene expression during Staphylococcus chromogenes subclinical mastitis. 2023 ADSA Annual Meeting. June 25-28, Ottawa, Canada.
- Mengqi Wang and Eveline M. Ibeagha-Awemu. Potential roles of DNA methylation in subclinical mastitis in Canadian Holstein cows. June 8th, 2023, Le cinquième webinaire Galactinnov. Online webinar.
- Mengqi Wang, Mario Laterrière, Pier-Luc Dudemaine, Nathalie Bissonnette, David Gagné, Jean-Philippe Roy, Marc-André Sirard, Eveline M. Ibeagha-Awemu. DNA methylation in first exon potentially regulate gene expression during bovine subclinical mastitis caused by Staphylococcus aureus. 2022 ADSA Annual Meeting. June 19-22, 2022, Kansas City, MO, USA.
- Mengqi Wang, Mario Laterrière, Pier-Luc Dudemaine, Nathalie Bissonnette, Xin Zhao, David Gagné, Eveline M. Ibeagha-Awemu. Integration of methylome and transcriptome profiles of milk somatic cells reveals involvement of DNA methylation in host immune response to bovine subclinical mastitis induced by Streptococcus uberis. World Congress on Genetics Applied to Livestock Production. July 3-8, 2022, Rotterdam, The Netherlands.
- Mengqi Wang, Nathalie Bissonnette, Mario Laterrière, Pier-Luc Dudemaine, David Gagné, Jean-Philippe Roy, Marc-André Sirard, Xin Zhao, Eveline M. Ibeagha-Awemu. Association of two DNA methylation haplotype blocks with health and production traits in Canadian Holstein cows. Meeting of the Dairy Cattle Breeding & Genetics Committee. October 4th 2022, University of Guelph, Guelph, ON, Canada.
- Mengqi Wang, Pier-Luc Dudemaine, Nathalie Bissonnette, Xin Zhao, Eveline M. Ibeagha-Awemu. Genome-wide DNA methylation profile of mammary gland tissues from Holstein cattle producing various milk protein yields. 2021 ASAS-CSAS-SSASAS annual meeting & trade show. July 14-17, 2021, Louisville Kentuchk, USA.
- Mengqi Wang, Pier-Luc Dudemaine, Nathalie Bissonnette, Eveline M. Ibeagha-Awemu. Role of DNA methylation in bovine mastitis. 2021 GRESABO summer symposium. 2021 September. Online webinar.
- Wang M., Ni W., Tang C., et al. Genome wide methylation analysis of mammary tissue with Staphylococcus aureus induced mastitis in Chinese Holstein cattle. The 19th National Symposium on animal genetics and breeding. October, 2018, Nanjing, Jiangsu, China. (Oral Presentation in Chinese).
- Wang M., Ni W., Guo J., et al. The Association Analysis between the Polymorphism of CXCR1 Gene Coding Region and Partial Reproductive Traits in Holstein Cattle. The 31st reproductive technology seminar of China Dairy Association. August, 2017, Hohhot, Inner Monggolia, China. (Oral Presentation in Chinese).
Poster presentations:
- Mengqi Wang, Nathalie Bissonnette, Mario Laterrière, David Gagné, Marc-André Sirard, and Eveline M. Ibeagha-Awemu. DNA Methylation alterations in repeat elements during subclinical mastitis caused by Staphylococcus aureus and Streptococcus uberis in Canadian Holstein cows. Plant and Animal Genome conference / PAG30. January 13-18, 2023, San Diego, USA.
- Mengqi Wang, Mario Laterrière, David Gagné, Nathalie Bissonnette, Marc-André Sirard, and Eveline M. Ibeagha-Awemu. DNA methylation alterations are potentially involved in cow’s response to subclinical mastitis due to Staphylococcus chromogenes. Plant and Animal Genome conference / PAG30. January 13-18, 2023, San Diego, USA.
- Mengqi Wang, Nathalie Bissonnette, Mario Laterrière, David Gagné, and Eveline M. Ibeagha-Awemu. DNA methylation alteration patterns in repeat elements are similar during subclinical mastitis caused by Staphylococcus chromogenes and Staphylococcus aureus. 39th International Society for Animal Genetics Conference. July 2-7, 2023, Cape town, South Africa.
- Mengqi Wang, Mario Laterrière, Pier-Luc Dudemaine, Nathalie Bissonnette, David Gagné, Jean-Philippe Roy, Marc-André Sirard, Eveline M. Ibeagha-Awemu. Identification of differential methylation regions as candidate discriminant markers for bovine subclinical mastitis caused by Staphylococcus aureus. 2022 ASAS-CSAS Annual Meeting & Trade Show. June 26-30, 2022, Oklahoma City, Oklahoma, USA.
- Mengqi Wang, Mario Laterrière, Pier-Luc Dudemaine, Nathalie Bissonnette, David Gagné, Eveline M. Ibeagha-Awemu. Potential involvement of DNA methylation of first intron in transcriptional regulation during bovine subclinical mastitis caused by Staphylococcus aureus. 2022 ASAS-CSAS Annual Meeting & Trade Show. June 26-30, 2022, Oklahoma City, Oklahoma, USA.
- Mengqi Wang, Mario Laterrière, Pier-Luc Dudemaine, Nathalie Bissonnette, David Gagné, Jean-Philippe Roy, Marc-André Sirard, Xin Zhao, Eveline M. Ibeagha-Awemu. Why epigenetics matters to dairy farmers – DNA methylation alterations are involved in bovine subclinical mastitis. Symposium sur les bovins laitiers. November 2, 2022, Drummondville, Québec, Canada.
- Mengqi Wang, Pier-Luc Dudemaine, Nathalie Bissonnette, Marc-André Sirard, Jean-Philippe Roy, Xin Zhao, David Gagné, Eveline M. Ibeagha-Awemu. Genome-wide DNA methylation alterations in milk somatic cells due to Streptococcus uberis subclinical mastitis. Le rendez-vous scientifique annuel d’Op+Lait 2021. Octobre 20, 2021,Centre des congrès de St-Hyacinthe, St-Hyacinthe, Québec, Canada.
- Mengqi Wang, Nathalie Bissonnette, Philip Griebel, Pier-Luc Dudemaine, Duy N. Do, Yongjiang Mao, Eveline M. Ibeagha-Awemu. Differentially expressed microRNAs with potential regulatory roles in ileum of Holstein cows with subclinical Johne’s disease. 2019 ASAS-CSAS Annual Meeting & Trade Show. July 8-11, 2019, Austin, Texas, USA.
- Mengqi Wang, Nathalie Bissonnette, Philip Griebel, Pier-Luc Dudemaine, Duy N. Do, Yongjiang Mao, Eveline M. Ibeagha-Awemu. Transcriptome analysis of ileal lymph nodes identifies key microRNAs affecting disease progression in Holstein cows with subclinical Johne’s disease. 2019 ASAS-CSAS Annual Meeting & Trade Show. July 8-11, 2019, Austin, Texas, USA.
Authorized patent
- Yongjiang Mao, Mengqi Wang, Wei Ni , Wenqiang Wang, Mingxun Li, Huimin Zhang, Zhangping Yang. A molecular marker and its detection method for auxiliary selection to reduce the susceptibility to leukemia in dairy cows. Patent No : ZL201710441975.1.



